Kinograte
kinograte.RmdIntroduction
Kinograte is used for network-based integration of multi-omics datasets. Kinograte is designed to utilize the prize-collecting Steiner forest (PCSF) algorithm. In this case, the goal of the PCSF algorithm is to identify a simplified sub network representative of a disease or a chemical perturbagen.
Example
Here, we present a simple example on how to use kinograte using three omic datasets (RNASeq, Proteomics, and Kinomics). The package comes bundled with datasets that be used to try out the package. These datasets will be used in this example.
Loading data
The input data for kinograte follow the format of the differential analysis output performed by standard analytic packages which typically consist of three main columns: (gene or protein symbol, log2 fold change (LFC), pvalue). We will use the gene or protein symbol as the feature identifier and either the LFC the pvalue as the metric to score and rank genes/proteins.
# rnaseq or micorarray example
reactable(rnaseq_example)Ranking and Standrizing Scores
First, we will percentile rank the features in each omic dataset:
# rank RNASeq based on LFC
rnaseq_example_ranked <- percentile_rank(rnaseq_example, `Gene name`, LFC)
reactable(rnaseq_example_ranked)Visualize the Ranked Features
You can visualize the ranked features with an interactive plot:
score_plot(rnaseq_example_ranked)or static figure:
score_plot(rnaseq_example_ranked, interactive = F)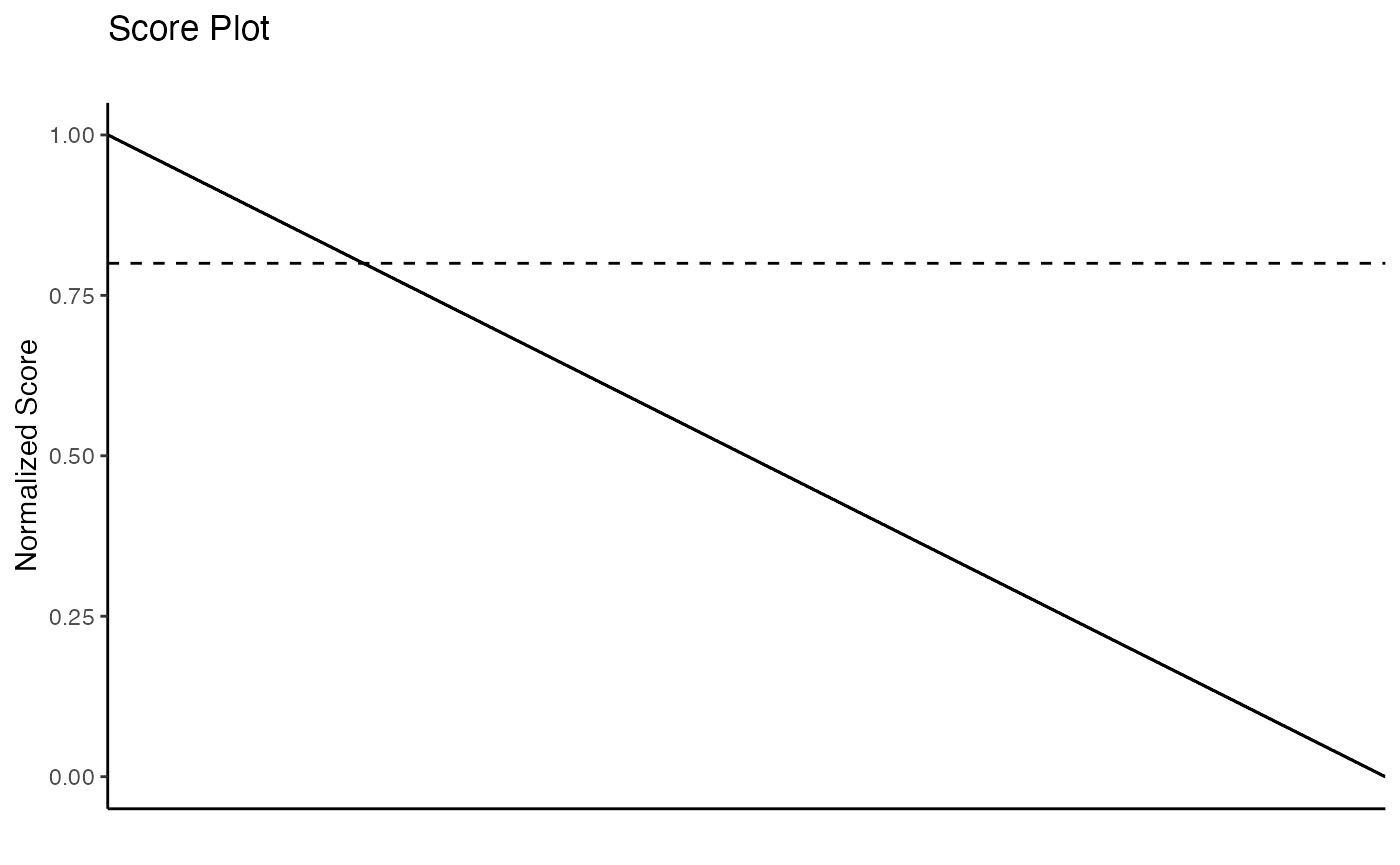
Extracting Top Hits
Next, we can extract the top hits from each omic dataset. We will be using the standrized scores as the main metric to set the threshold (the threshold can be adjusted):
rnaseq_example_top <- top_hits(rnaseq_example_ranked, prec_cutoff = 0.95, omic_type = "RNA")
reactable(rnaseq_example_top)Combining Top Ranked Features
Next, we will combine the top ranked features from each omic dataset. This combined dataframe will be used the input the network-based integration function:
combined_df <- combine_scores(rnaseq_example_top, proteomics_exmaple_top, kinomics_exmaple_top)Multi-omics Integration
We will use the combined dataframe to integrate the omics datasets utilizing the PCSF algorithm to generate an integrated biological network and using the built-in protein-protein interaction (PPI) dataset:
# note: the ppi_network can be replcaed with a user-provided ppi network file
kinograte_res <- kinograte(combined_df, ppi_network = ppi_network_example, cluster = T)
#> 90.57% of your terminal nodes are included in the interactome
#> Solving the PCSF by adding random noise to the edge costs...Multi-omics Integration Visualization
To visualize the integrated network, we will use the visualize_network function:
visualize_network(nodes = kinograte_res$nodes, edges = kinograte_res$edges,
cluster_df = kinograte_res$wc_df, options_by = NULL)You can also add more options for the dropdown menu:
Group by omic type
visualize_network(nodes = kinograte_res$nodes, edges = kinograte_res$edges,
cluster_df = kinograte_res$wc_df, options_by = "group")Group by cluster
visualize_network(nodes = kinograte_res$nodes, edges = kinograte_res$edges,
cluster_df = kinograte_res$wc_df, options_by = "cluster")
enrichr_res <- network_enrichment(kinograte_res$network)
#> Performing enrichment analysis...
#>
#> Enrichment is being performed by EnrichR (http://amp.pharm.mssm.edu/Enrichr) API ...
reactable(enrichr_res$pathways)